Hypothetical tonnages generated by seeded randomizer. GHG emission rates will be sourced from EPA’s WARM v. 15 for food waste in short tons (to correct to metric).
Tonnages by Location
| TreatmentTYpe | Emission (CO2equiv/t Food Waste) | Facility ID | Food Waste (t/year) |
|---|---|---|---|
| TreatmentTYpe | Emission (CO2equiv/t Food Waste) | Facility ID | Food Waste (t/year) |
| AD-leachate | -0.02 | 9 | 1277.6667 |
| AD-SSO | -0.04 | 13 | 235.6667 |
| AD-SSO | -0.04 | 14 | 7997.6667 |
| Incineration | -0.13 | 6 | 7949.4944 |
| Incineration | -0.13 | 7 | 2219.7040 |
| Incineration | -0.13 | 8 | 1454.4256 |
| Compost | -0.12 | 10 | 38253.0000 |
| Compost | -0.12 | 11 | 2200.3333 |
| Compost | -0.12 | 12 | 53311.0000 |
| Landfill | 0.50 | 3 | 0.0000 |
| Landfill | 0.50 | 4 | 48.2480 |
| Landfill | 0.50 | 5 | 1982.2824 |
Mind-mapping/Flowcharting the paths using DiagrammeR
Source separated food waste paths
Mixed garbage paths
Where are the facilities relative to the transfer stations? Will some transportation emissions associated with these facilities be disproportionate due to haulage distance?
Reading layer `YR-FoodWaste' from data source `C:\Users\HoJa\Documents\R\passerium\data\YR-FoodWasteADaltered.kml' using driver `KML'
Simple feature collection with 14 features and 2 fields
geometry type: POINT
dimension: XYZ
bbox: xmin: 74.99062 ymin: 42.02855 xmax: 82.56345 ymax: 45.3053
epsg (SRID): 4326
proj4string: +proj=longlat +datum=WGS84 +no_defsMapping it out >>
preparing for next stage
#create edge & node join for plot based on r-bloggers 2018 method #https://www.r-bloggers.com/2018/05/three-ways-of-visualizing-a-graph-on-a-map/
#making connectors
locs_lat_lon<-dplyr::select(locs_lat_lon, index, X,Y, Name)
edges_for_plot <- links %>%
inner_join(locs_lat_lon %>% dplyr::select(index, X, Y), by = c('source' = 'index')) %>%
rename(x = X, y = Y) %>%
inner_join(locs_lat_lon %>% dplyr::select(index, X,Y), by = c('target' = 'index')) %>%
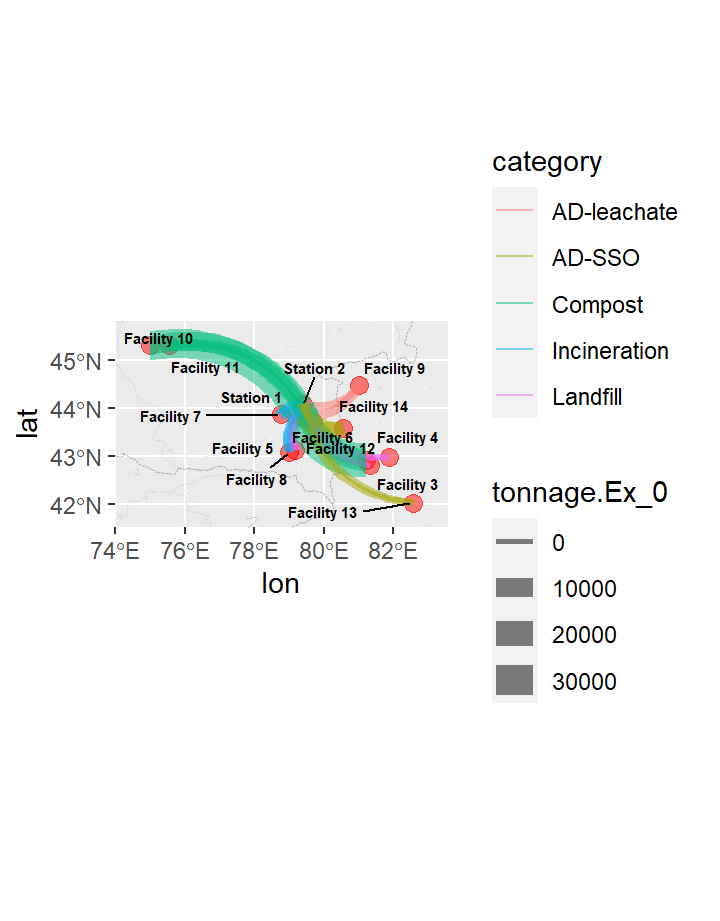
rename(xend = X, yend = Y)Showing where the facilities are and trying to demarcate type of process undertaken there, as well as sizing markers by annual tonnages received.
Then, create edges & nodes join to ready for connectors.
Food waste-hurling fun - Creating weighted arrows showing flows from transfer stations (1,2) to the facilities (3 through 14)
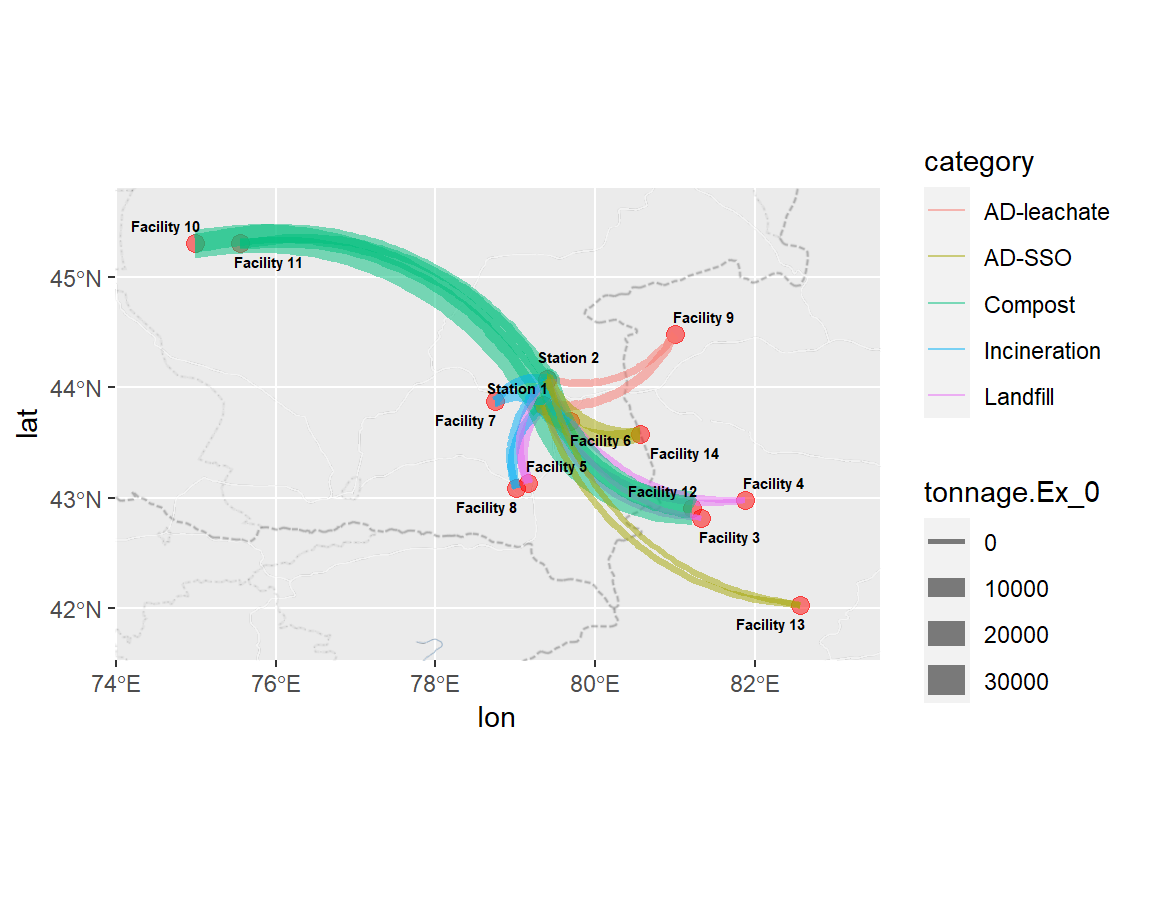
TBD - mini bar charts at each location showing treatment emissions vs. transport